Documentation of the manufacturing process
During biologics development, including upscaling and changes to the manufacturing process, you must demonstrate clearance of process-related impurities from the product. Even in trace amounts, residual impurities, such as Host Cell Proteins (HCPs), can affect product quality, efficacy, and safety. HCPs qualify as a critical quality attribute (CQA) in biologics production – and authorities demand that you monitor them closely.
Mass spectrometry (LC-MS) analysis provides detailed documentation of HCP levels across process samples, and the resulting data provides a valuable tool for designing and evaluating HCP purification strategies. Mass spectrometry information about each HCP identity and quantity enables data-driven removal rather than trial and error experiments. E.g., you can make efficient removal steps for the individual HCP based on attributes like pI and molecular weight.
With MS data, you have solid documentation of removing problematic HCPs and documenting their clearance and control strategy for regulatory license applications.
Uses for LC-MS in process optimization
Troubleshooting & bridging studies
Identify and quantify HCPs and compare different downstream purification strategies for HCP clearance
Comparability testing
Monitor HCP levels during process changes, scale-up, and tech-transfer batches between CMOs
Process Performance Qualification (PPQ) analysis
Demonstrate the consistent clearance of HCPs across multiple GMP PPQ runs
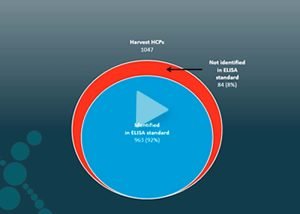
Downstream development is often a matter of trying different purification steps and evaluating the effect of each. Matrix experiments take time, and the success rate may depend on complex matrix experiments or pure luck. Analysis using a standard ELISA only provides a single number representing an estimate for the total HCP amount and does not provide details of the individual amounts.
On the contrary, mass spectrometry analysis provides each protein's identity, quantity, and attributes. Based on the data, you can evaluate the efficiency of current removal steps or design new steps targeting specific HCPs.
MS data can also help increase the yield of the Drug Substance (in some cases 10-20%) by optimization of purification procedures (columns, pH, gradients, fraction-collection), thereby bringing down the cost of produced material (COGS) while monitoring HCPs to ensure that process changes do not increase the content of critical HCPs.
You may already have a custom ELISA developed for your process but then decide to shift CMOs or re-design the purification process. Such changes may affect the ELISA's performance or coverage since the HCP profile may have changed. Often, this results in 1-2 years for developing a new set of anti-HCP antibodies and validating the assay.
Instead, you could consider using LC-MS as an orthogonal method to support your strategy. The current ELISA might still be fit-for-purpose if you can document the changes in the process and ELISA coverage. The method can be used for detailed characterization of the ELISA reagents and to evaluate the coverage of individual HCPs before and after process changes. We can also set up an MS assay to quantify HCP levels for you within weeks. Thereby, you save valuable project time and costs. And in case your HCP ELISA fails, the MS assay may even be validated according to ICH guidelines and used for GMP release testing.
Process performance qualification (PPQ) is essential to process validation during or after clinical phase III manufacturing. With the PPQ runs, you must show that your process can consistently deliver a quality product and document that the HCP level in the end product is consistent. As authorities increasingly ask for detailed data by orthogonal methods to ELISA, i.e., MS, why not include detailed data on individual HCPs clearance across the three PPQ runs?
For each step in the purification process, from the early harvest sample through the different purification steps to the pure drug substance, mass spectrometry provides the quantity of each HCP and shows how they are cleared. Comparing the HCP clearance across PPQ runs gives the complete picture and control of process consistency. Furthermore, the data can form part of regulatory documentation in the Biologics License Application (BLA) for the regulatory agency.
Typical project process
You typically work with
these experts:


Project scope
We like to start with an online meeting to learn more about your project. Based on your needs, you will receive a draft proposal outlining the suggested analyses and expected timeframe.
Samples
After signing the final project proposal, we will contact you for details about your samples. We will inform you of an estimated report delivery date as soon as we receive your samples.
Execution
A project leader will oversee the project and send you status updates by email at regular intervals.
The analysis varies according to the project but typically includes:
- An optimized sample preparation developed explicitly for HCP samples.
- Analysis using a robust microflow HPLC and SWATH® MS/MS system to ensure high reproducibility.
- A database for protein identification specifically for your expression system and drug substance.
- Spike-in of internal standard proteins (0-2000 ppm), enabling quantification linearity even to low ppm levels.
Results
You will receive the report by email. As standard, the analysis report includes:
- Objectives, description of analytical procedure, results, and conclusions.
- List of the total HCPs in the sample.
- List of individual HCPs and their quantities.
- Evaluation of HCPs of potential concern.
- Selected raw data, e.g., excel sheets. You may request additional raw data if needed.
Follow up
Upon completion of the project, your team is invited to review the results at an online meeting.
Curious to know more?

Whatever challenge or question you may have, we are here to help you solve it. One of our protein analysis experts will discuss the best analysis approach or method for your minimizing HCP levels in your project by email or online meeting – without obligation.
Client stories
Video: What is causing polysorbate degradation?
Video: Can your CDMO tackle Host Cell Proteins?
Video: Mock and harvest HCP comparison
Analysis of individual human residual proteins
Video: Orthogonal HCPs of mAb product
Video: HCP purification optimization from harvest to final DS
Video: Comparison of HCP removal strategies
Purified mAb with difficult-to-remove HCP impurity (ubiquitin)
Webinar: Data-driven process development using HCP analysis by LC-MS
White paper: HCP analysis of downstream process samples by LC-MS
Documenting product quality and process consistency
Orthogonal HCP analysis for vaccine development
Benefits of applying LC-MS in HCP process optimization
Early detection of HCPs of concern
Identify and quantify
process-related
impurities for
risk assessment
Data-driven process optimization
Compare downstream purification strategies
or create one based on HCP attributes
Faster process development
Evaluate if a
commercial
ELISA provides
sufficient coverage
Monitor process Scale-Up
Monitor HCP levels
during scale-up and
tech-transfer
between CMOs
Document Process Performance (PPQ)
Qualify and document production robustness,
consistency, and replicability
Benefits of applying LC-MS in process optimization
Early detection of HCPs of concern
Identify and quantify
process-related
impurities for
risk assessment
Data-driven purification optimization
Compare downstream purification strategies
or create one based on HCP attributes
Faster process development
Evaluate if a
commercial
ELISA provides
sufficient coverage
Monitor process Scale-Up
Monitor HCP levels during scale-up and
tech-transfer
between CMOs
Process Performance Qualification (PPQ)
Document production robustness,
consistency, and replicability
What clients say
Knowledge center
Why use intact standards to normalize MS data when quantifying HCPs instead of heavy-labeled peptides?
Intact proteins added to the sample undergo the same treatment as the sample proteins. The denaturing conditions, digest conditions, and matrix effects are the same for all proteins in the mix, and we can use the added proteins as trustworthy internal calibration standards.
Heavy-labeled peptides are not affected by the sample preparation in the same way and do not account for the effects of the treatment on the sample proteins. For this reason, heavy-labeled peptides are not our preferred choice.
Is it possible to follow the clearance of a specific HCP in the downstream process?
Analyzing a range of in-process samples using MS makes it possible to compare the sample's ppm levels of an individual HCP through purification steps. The data enables your process development team to adjust the manufacturing to remove specific HCPs from the product entirely.
Research article:
Videos:
- Analysis of gene therapies
- Analysis of residual protein in viral products
- Evaluating purification strategies
- When no commercial ELISA kit fits your expression system
- Analysis of bacteriophage product
- Residual biocatalysis in API
- Non-dilutional linearity of ELISA: Hitchhiker HCPs
- Optimized analysis of peptides for clinical trials
Poster:
Cases:
- Analysis of individual human residual proteins using an orthogonal method
- Documenting product quality & process consistency by analysis of residuals
- Orthogonal HCP analysis for mAbs
Protocol:
Talk to us
Whatever protein-related challenge or question you may have, we would love to help. Our experts can help you decide on the best analytical approach for your project by email or online meeting - providing advice without obligation.