Literature
Why you need accurate concentration determination of protein standards
January 18 2017, by Thomas Kofoed

Question:
How do I quantify my protein standard?
How do I quantify my protein standard? Is it true that if done incorrectly, it can affect the determination of my protein concentration?
Answer:
Have you ever questioned the quantitative data you get from Bradford, Lowry, or BCA protein quantification assays? These traditional methods are speedy and can be performed in most laboratories. They are based on UV absorption (A280) of the protein relative to a standard, typically BSA, of known concentration [1, 2].
- Unfortunately, these assays are prone to interference from chemicals and the presence of other proteins.
- Furthermore, your purified protein may have a different UV absorption coefficient than the BSA calibration standard.
Therefore the concentration determination of the protein standard can be wrong! All your experiments based upon the amount in the protein reference standard could also be wrong! Imagine this impact on your assay or your downstream process development [1, 2].
This is why you should use triplicate amino acid analysis (AAA) to quantify your protein standard [3] accurately.
Amino acid analysis – the gold standard for protein concentration determination
There are several good reasons why you should use triplicate AAA [2] when you need quantitative data you can trust – for instance, to analyze your protein standard solutions:
- It is a very robust analysis. It can be performed in the presence of salt and detergents, as it is based on ion-exchange chromatography. The only contaminant obstructing the analysis is a chemical containing primary amines. And even then, it might still be possible to do an accurate quantification if the calculations can be based on amino acids not affected by the contaminant.
- You can compare the results with the sequence. This is an essential feature of the analysis. By comparing the quantitative data for each amino acid residue with the amino acid sequence of the protein, you will get information about the purity of your sample. If the data does not fit within a few residues, your sample is most likely contaminated by other proteins.
- The analysis can measure from 100-10.000 pmol with high linearity. It is linear over a broad concentration range. Therefore, it is suitable for samples with an unknown protein concentration.
- Your data can be qualified using internal and external reference standards. You obtain the most accurate result by analyzing each sample in triplicate. This means that each sample is analyzed as three individual samples from hydrolysis through data calculation. You use standard protein solutions in each batch analysis to determine the test’s precision and accuracy. Adding an internal standard amino acid (e.g., Sarcosine or Norleucine) to each sample makes it possible to correct changes in reagent stability and HPLC flow rate [1-3].
How do we approach protein concentration determination at Alphalyse?
At Alphalyse, we quantify sixteen amino acids. These are the standard 20 amino acids, except tryptophan and cysteine, which are degraded by hydrolysis. We thus determine Asparagine as aspartic acid and glutamine as glutamic acid. I always suggest running each sample in triplicate, and we also include two BSA samples (NIST standard) to determine the precision and the accuracy of each batch analysis.
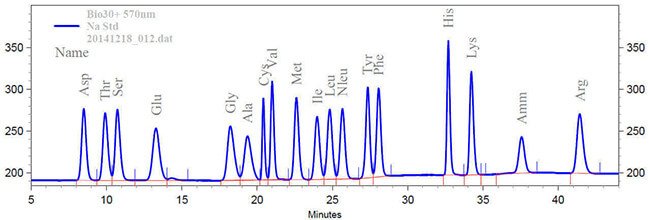
Figure: HPLC separation of amino acids for identification and quantification
To calculate the data, you first take the measured AA amount in pmol divided by the number of each specific amino acid in the sequence providing a pmol protein. Then you calculate the average of these numbers in pmol using the amount of protein in µg and the molecular weight of the protein.
Finally, we perform the calculations using the best linear fit based on detected amino acids. We modify each experiment to avoid including amino acids with greater than 5 percent variation.
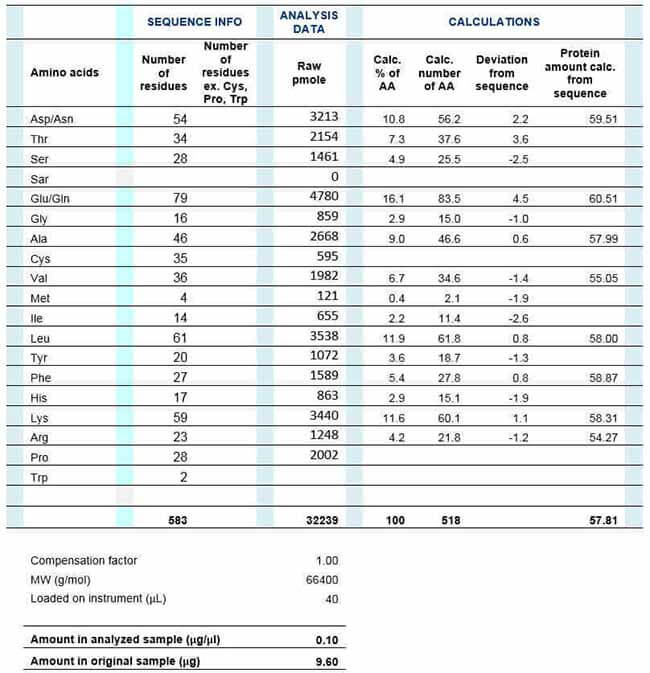
Table: Calculating the accurate amount of protein
To read more about our triplicate AAA service, please go to this site
References
[1] Rutherfurd et al.: “Amino acid analysis,” Current Protocols in Protein Science, 2009
[2] Rutherfurd et al.: “Quantitative amino acid analysis,” Current Protocols in Protein Science, 2011
[3] Noble et al.: “Quantification of protein,” Methods in Enzymology, 2009
Talk to us
Whatever protein-related challenge or question you may have, we would love to help. Our experts can help you decide on the best analytical approach for your project by email or online meeting - providing advice without obligation.