Literature
Question:
How can I improve label-free quantification of HCP?
I find it difficult to get an accurate label-free quantification of Host Cell Proteins with traditional multiple reaction monitoring using LC-MS. It also seems that my data is difficult to reproduce. – Even if I perform multiple LC-MS runs for each sample.
Is there anything I can do to improve on the quantification and the reproducibility of the data?
Answer:
For quantitative proteomics experiments, it has been a normal procedure to include an isotope-labeled peptide standard to obtain good HCP data [1].
Another solution is using chemical labeling (like iTRAQ® or SILAC) [2]. Some people even use isotope labeled recombinant proteins. You do this to consider the digestion efficiency of a full-length protein [3].
All of the above methods are time-consuming – and they are not always possible to use [2].
So what can you do instead?
With the new SWATH® LC-MS technology, label-free quantification of HCPs is no longer a problem
The reasons for this are:
- SWATH® analysis is a data-independent acquisition method. This results in very reproducible MS data – both for the identification and quantification of HCPs [4].
- Accurate absolute quantification of HCPs is now obtained by adding standard proteins. They are added in known amounts and are digested within the sample.
- The SWATH® LC-MS is running at robust microflow rates. Therefore, there is no longer a problem with time-consuming nanoflow HPLC or carry-over.
Label-free quantification of thousands of proteins
In order to test the new LC-MS method, we applied SWATH® label-free quantification to an E. coli cell lysate. With this method, we could identify and quantify more than 1.000 proteins, up to a total sum of 969.158 ppm. The expected sum is 1.000.000 ppm – see graph below.
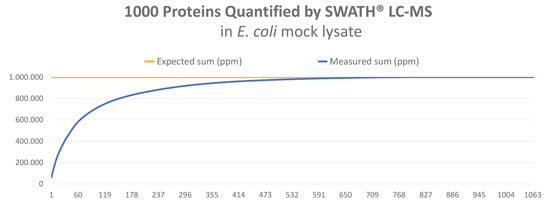
The analysis is so robust and reproducible that the standard deviation for individual proteins is below 20% in repeat experiments.
So to sum it up:
With the SWATH® technique it is now much easier to quantify proteins reproducibly. And in addition, it is done without isotope labeling.
More information on analysis of HCP:
The SWATH® technology is a form of data-independent analysis (DIA). You can learn more about the technology here: “What is SWATH”
Find out more on the identification and quantification by Host Cell Protein analysis on the Alphalyse website
References
[1] Zhu-Shimoni et al: “Host Cell Protein Testing by ELISAs and the Use of Orthogonal Methods“, Biotechnology and Bioengineering, 2014
[2] Tscheliessnig et al: “Host cell protein analysis in therapeutic protein bioprocessing – methods and applications.“, Biotechnology Journal, 2013
[3] Bracewell et al: “The Future of Host Cell Protein (HCP) Identification During Process Development and Manufacturing Linked to a Risk-Based Management for Their Control“, Biotechnology and Bioengineering, 2015
[4] Heissel et al: “Evaluation of spectral libraries and sample preparation for DIA-LC-MS analysis of host cell proteins: A case study of a bacterially expressed recombinant biopharmaceutical protein.“, Protein Expression and Purification, 2018
Talk to us
Whatever protein-related challenge or question you may have, we would love to help. Our experts can help you decide on the best analytical approach for your project by email or online meeting - providing advice without obligation.
