Literature
Question:
Should we worry about ubiquitin in our mAb?
Our process-specific HCP ELISA did not detect Host Cell Proteins (HCPs) in our purified monoclonal antibody (mAb). However, mass spectrometry-based HCP analysis identified ubiquitin in low-ppm amounts.
Do you have any suggestions for why the Protein A purification did not remove it? Why did ELISA not detect it? And should we worry about its presence?
Answer:
We saw the same problem in a recent client project and also observed ubiquitin in analyses of other mAb products. Therefore, it seems that it has several characteristics that makes it difficult to remove by regular mAb purification steps.
Before discussing these aspects, I would like to explain the characteristics and implications of ubiquitin. I.e., if you should worry about it altering your product or even harming the patient.
Ubiquitin – problematic or harmless?
Ubiquitin is a small regulatory protein of 76 amino acids. It alters protein fate by changing stability, function, subcellular localization, and interactions with other proteins [1-5]. When it is covalently attached to protein substrates, a process known as ubiquitination, it targets degradation by the ubiquitin-proteasome system. This post-translational modification is essential for cellular homeostasis.
With these abilities, it may seem unfavorable to have ubiquitin in the drug product. Nonetheless, no one has yet documented that it can cause immunogenic reactions in patients or affect drug stability.
You can investigate if ubiquitin is present in your samples and monitor its levels between batches with mass spectrometry data. Then – if you find it necessary – you can remove it based on its physicochemical properties.
Why was ubiquitin not removed during purification?
It is difficult to say why the purification process did not remove the ubiquitin. Perhaps it was covalently bound to the drug substance as a low-level post-translational modification. Or maybe it was just tightly associating with the mAb? It may happen since some HCPs attach non-covalently to the drug molecule as so-called hitchhiker HCPs. Alternatively, it may co-purify with the mAb if their physiochemical properties are similar. However, ubiquitin is a small protein of 8 kDa, whereas the mAb is 150 kDa. So this is not likely the problem.
Across the many mAbs analyzed by Alphalyse, we often identify proteins from the ubiquitin-related pathway. And the data indicate that ubiquitin binds tightly, covalently, or non-covalently to the mAb. This experience further supports that the otherwise efficient Protein A step does not remove ubiquitin as efficiently as other HCPs.
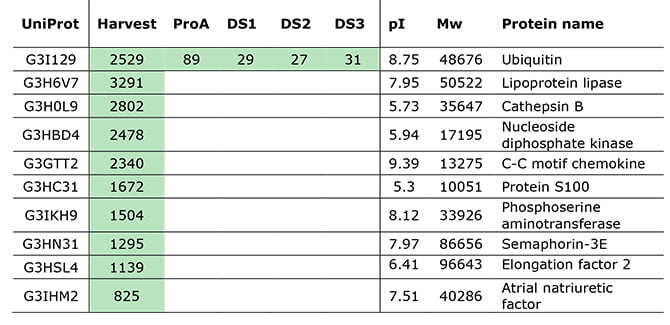
Why did the process-specific ELISA not detect ubiquitin?
In our client’s purified mAb, mass spectrometry (MS) determined the samples’ ubiquitin to be 1-25 ppm. The ubiquitin residual was thus within the detectable range of the ELISA. However, its relatively small size may be why ELISA missed it. ELISAs often have trouble identifying low Mw HCPs, since small Mw proteins are poor immunogens. I.e., they are not raising any high-specificity antibodies for the HCP ELISA [6].
A simple BLAST in UniProtKB reveals more than 250 variants from multiple organisms with above 90% sequence similarity. Furthermore, high homology could also play a role since ubiquitin conserves highly across species. This fact could make the CHO ubiquitin non-immunogenic in the rabbit used for raising the ELISA antibodies.
An analysis that detects low-ppm HCPs
To investigate the problem further, you may turn to mass spectrometry analysis.
Specifically, for mAb products, Alphalyse utilizes a highly sensitive method, which uses native digest to lower the detection limit.
You can learn more about the HCP analysis for monoclonal antibodies on the Alphalyse website
References
[1] Goldstein et al.: “Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells,” Proceedings of the National Academy of Sciences of the United States of America, 1975
[2] Wilkinson, KD: “The discovery of ubiquitin-dependent proteolysis,” Proceedings of the National Academy of Sciences of the United States of America, 2005
[3] Kimura et al.: “Regulatory mechanisms involved in the control of ubiquitin homeostasis,” Journal of Biochemistry, 2010
[4] Glickman et al.: “The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction,” Physiological Reviews, 2002
[5] Lin et al.: “Exploitation of the host cell ubiquitin machinery by microbial effector proteins,” Journal of Cell Science, 2017
[6] Bracewell et al.: “The Future of Host Cell Protein (HCP) Identification During Process Development and Manufacturing Linked to a Risk-Based Management for Their Control,” Biotechnology and Bioengineering, 2015
Talk to us
Whatever protein-related challenge or question you may have, we would love to help. Our experts can help you decide on the best analytical approach for your project by email or online meeting - providing advice without obligation.
